Our research focuses on functional and evolutionary aspects of small RNA pathways. This involves the development of
bioinformatics tools for analysis of high-throughput sequencing data to characterize different classes of small RNAs.
Background: Jumping Genes show Background: Jumping Genes hide
 In
1950 Barbara McClintock first described mobile DNA elements in maize, where transposing DNA elements cause reversible disruptions of coloring
genes (right top: Science cover from 20. Nov. 2009, Photo: Kate Mathis/Getty Images). Though McClintocks ideas were initially
considered sceptically, she later on received great honors including the Nobel Prize for Physiology and Medicine 1983 for her pioneering
work. Today, diverse types of transposable elements are described in almost all studied eukaryotic species. Transposable elements are typically
assorted into class 1 and class 2 transposons. While the former propagate via RNA intermediaries that are subject to reverse
transcription, class 2 transposons are excised from one locus and re-integrated at a different site via the enzyme
In
1950 Barbara McClintock first described mobile DNA elements in maize, where transposing DNA elements cause reversible disruptions of coloring
genes (right top: Science cover from 20. Nov. 2009, Photo: Kate Mathis/Getty Images). Though McClintocks ideas were initially
considered sceptically, she later on received great honors including the Nobel Prize for Physiology and Medicine 1983 for her pioneering
work. Today, diverse types of transposable elements are described in almost all studied eukaryotic species. Transposable elements are typically
assorted into class 1 and class 2 transposons. While the former propagate via RNA intermediaries that are subject to reverse
transcription, class 2 transposons are excised from one locus and re-integrated at a different site via the enzyme
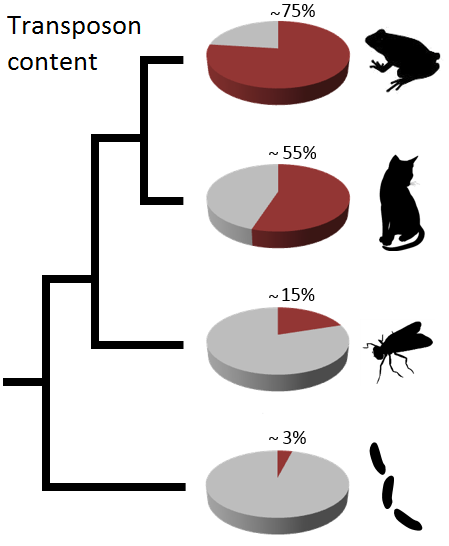 transposase. Mobile DNA can account for a considerable fraction of metazoan genomes. Conservative estimations assume that roughly 50% of a mammalian genome
are made up of repetitive DNA. Especially class 1 transposons of endogenous (e.g. the human specific 7SL RNA-derived Alu elements) or exogenous (e.g. ERV)
origin steadily increased in copy number by applying the copy-paste-like cloning mechanism for their own propagation throughout the host genome,
resulting in an increase of copy number with each transposition.
transposase. Mobile DNA can account for a considerable fraction of metazoan genomes. Conservative estimations assume that roughly 50% of a mammalian genome
are made up of repetitive DNA. Especially class 1 transposons of endogenous (e.g. the human specific 7SL RNA-derived Alu elements) or exogenous (e.g. ERV)
origin steadily increased in copy number by applying the copy-paste-like cloning mechanism for their own propagation throughout the host genome,
resulting in an increase of copy number with each transposition.
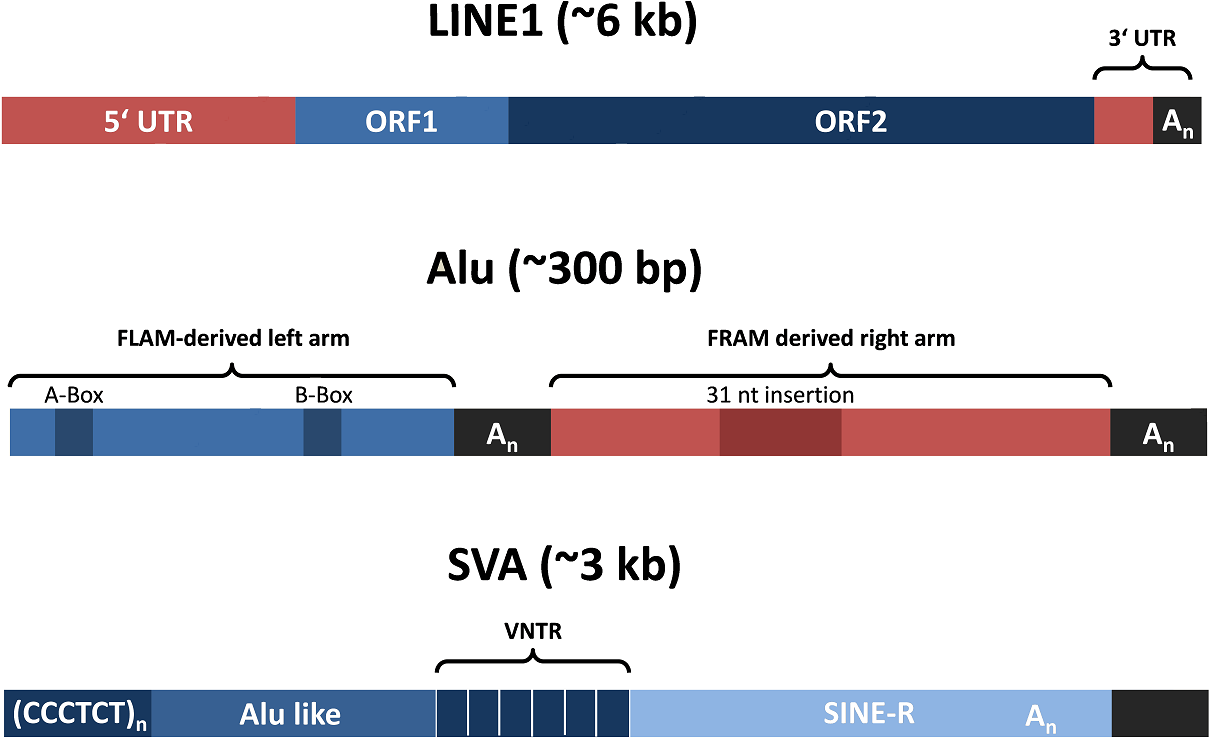 Different classes of transposons are active in diverse animals and constitute a steady threat to genome integrity by potentially disrupting essential genes and providing the prerequisite for ectopic
recombination (right bottom: Active human class 1 transposons). Throughout the metazoan lineage, organisms have established a molecular defense mechanism that employes Piwi proteins
and a broad arsenal of small noncoding RNAs (Piwi-interacting RNAs, piRNAs) to track down hostile retroposon RNA-intermediates via RNA interference (RNAi). On the other
hand, new classes of retroposons arised and existing retroposons constantly diversified over evolutionary timespans thus eluding the established host resistence. Constant host adaptation results in a
molecular arms race between transposons and the Piwi/piRNA pathway components resulting in fast evolutionary changes driven by positive selection. Evolutionary
patterns become even more complex considering benefical functionalization events of transposon sequences through e.g. exonization, promotor capturing or miRNA processing from terminal
inverted repeats (TIRs) of class 2 elements.
Different classes of transposons are active in diverse animals and constitute a steady threat to genome integrity by potentially disrupting essential genes and providing the prerequisite for ectopic
recombination (right bottom: Active human class 1 transposons). Throughout the metazoan lineage, organisms have established a molecular defense mechanism that employes Piwi proteins
and a broad arsenal of small noncoding RNAs (Piwi-interacting RNAs, piRNAs) to track down hostile retroposon RNA-intermediates via RNA interference (RNAi). On the other
hand, new classes of retroposons arised and existing retroposons constantly diversified over evolutionary timespans thus eluding the established host resistence. Constant host adaptation results in a
molecular arms race between transposons and the Piwi/piRNA pathway components resulting in fast evolutionary changes driven by positive selection. Evolutionary
patterns become even more complex considering benefical functionalization events of transposon sequences through e.g. exonization, promotor capturing or miRNA processing from terminal
inverted repeats (TIRs) of class 2 elements.
A non-representative selection of relevant literature
Biémont and Vieira 2006 Nature 443:521-524 PubMed link
Slotkin and Martienssen 2007 Nat Rev Genet 8:272-285 PubMed link
Hancks and Kazazian 2012 Cur Opin Genet Dev 22:191-203 PubMed link
Fedoroff 2012 Science 338:758-767 PubMed link
Lisch 2013 Nat Rev Genet 14:49-61 PubMed link
Background: Small RNAs show
Background: Small RNAs hide
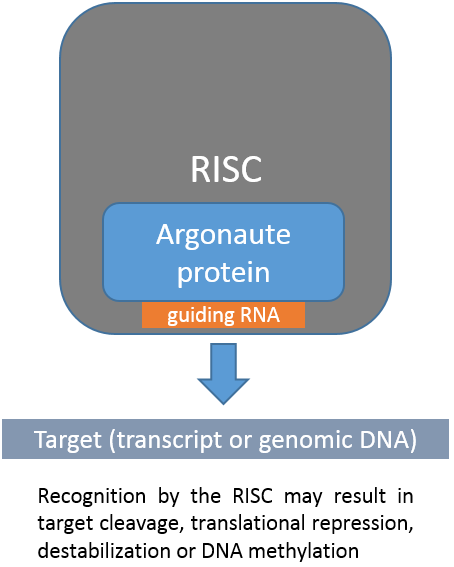
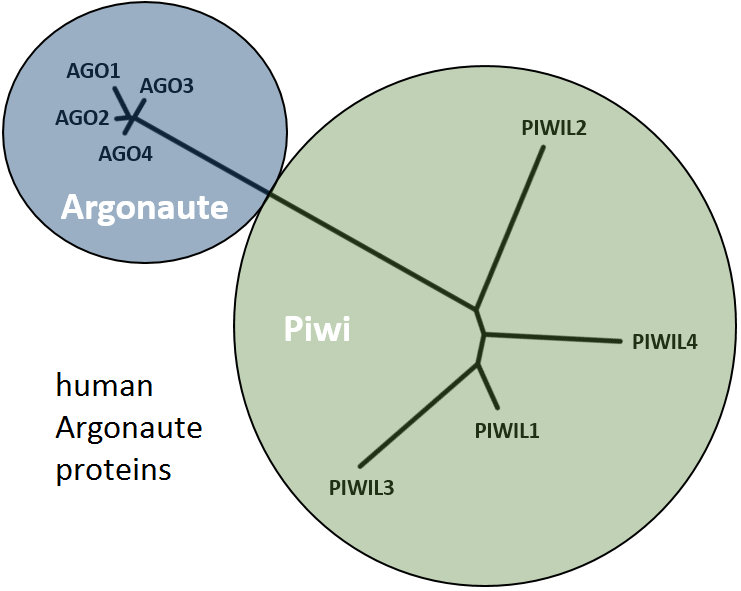 Small noncoding RNAs (sncRNAs) represent a diverse class of regulatory RNAs ranging from 18-34 nt in length being involved in a variety of
biological processes. Bound to Argonaute proteins they act at the heart of RNA induced silencing complexes (RISCs) to regulate gene expression
or defend the genome from transposable elements on the transcriptional and post-transcriptional level.
Small noncoding RNAs (sncRNAs) represent a diverse class of regulatory RNAs ranging from 18-34 nt in length being involved in a variety of
biological processes. Bound to Argonaute proteins they act at the heart of RNA induced silencing complexes (RISCs) to regulate gene expression
or defend the genome from transposable elements on the transcriptional and post-transcriptional level.
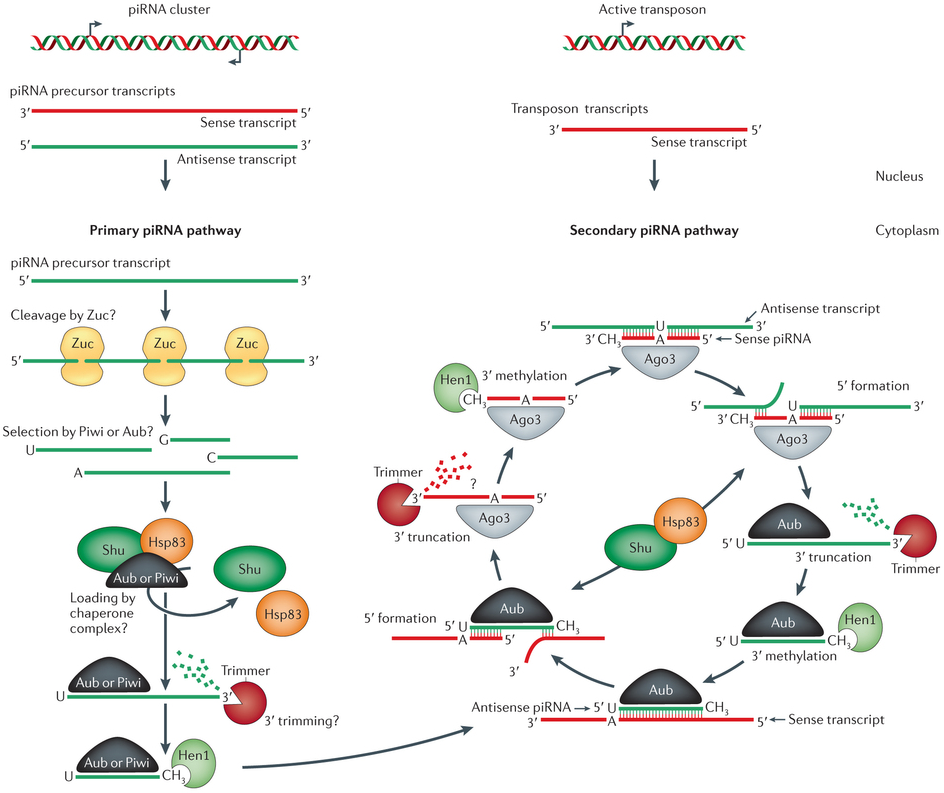
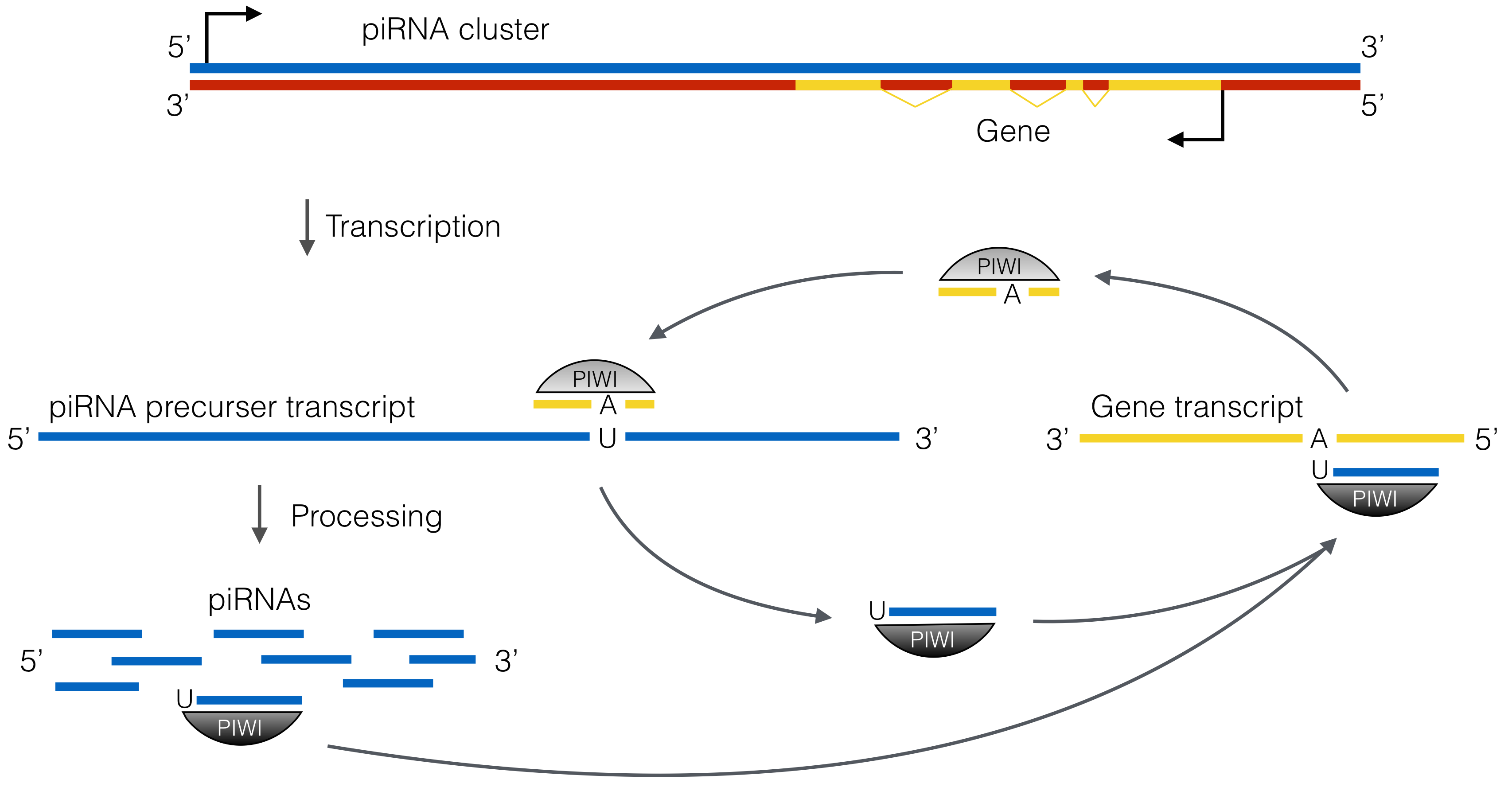
sncRNAs can be categorized into three main classes (further classes exist): micro (mi-) RNAs, short interferring (si-) RNAs and Piwi-interacting (pi-) RNAs. Both, miRNAs and siRNAs are products of Dicer processing, an enzyme that recognizes double stranded RNA, and bind to the Argonaute-like subclass of Argonaute proteins. In contrast, piRNAs are processed from single stranded transcripts either by Zucchini (primary piRNAs from piRNA cluster transcripts) or in a self sustaining amplification loop referred to as ping-pong-cycle involving typically two different paralogs of the Piwi-like subclass of Argonaute proteins that alternately slice transcripts from piRNA clusters and complementary target loci such as (but not exclusively) transposons (bottom: piRNA biogenesis. Figure from Luteijn and Ketting 2013. Nature Review Genetics 14:523-534 with reuse liscence #3595390394990).
 While miRNAs are involved in post-transcriptional gene regulation in both plants and animals, piRNAs and Piwi proteins (that are absent in
plants) have been linked to transciptional and post-transcriptional transposon control in the animal germline, though evidence for diverse functions beyond
transposon repression is mounting. In plants, transposons are controlled by siRNAs which also represent the major defense line against RNA viruses. In
contrast, animal siRNAs are involved in transposon controll and gene regulation though the significance of siRNAi in animals remains an
outstanding question and appears to be relegated to the background compared to miRNA- and piRNA-pathways.
While miRNAs are involved in post-transcriptional gene regulation in both plants and animals, piRNAs and Piwi proteins (that are absent in
plants) have been linked to transciptional and post-transcriptional transposon control in the animal germline, though evidence for diverse functions beyond
transposon repression is mounting. In plants, transposons are controlled by siRNAs which also represent the major defense line against RNA viruses. In
contrast, animal siRNAs are involved in transposon controll and gene regulation though the significance of siRNAi in animals remains an
outstanding question and appears to be relegated to the background compared to miRNA- and piRNA-pathways.
- How secondary structure influences small RNA pathways
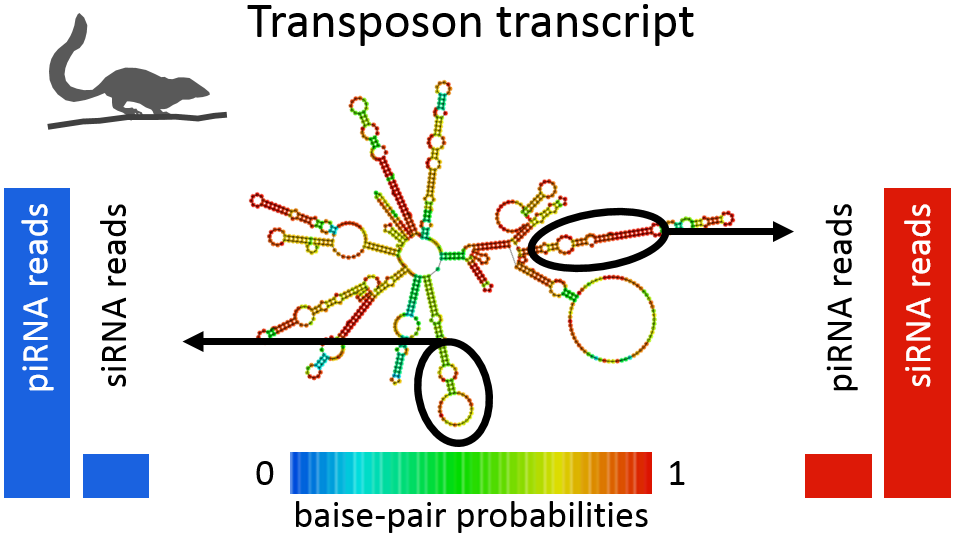
Transposable elements in the tree shrew are targeted not only by piRNAs but also by smaller RNAs resembling typical features of siRNAs. Comprehensive bioinformatic analyses showed that piRNAs preferentially target transposon transcripts at sites that do not form foldback structures and thus remain accessible for guiding piRNAs. In contrast, sites that avoid piRNA targeting owing to strong secundary structures are processed by Dicer, an enzyme that recognizes double-stranded RNA (Rosenkranz et al. 2015, RNA 21(5):911-922).
Background: Jumping Genes show Background: Jumping Genes hide
__________________________________________________________________________________________________________
 In
1950 Barbara McClintock first described mobile DNA elements in maize, where transposing DNA elements cause reversible disruptions of coloring
genes (right top: Science cover from 20. Nov. 2009, Photo: Kate Mathis/Getty Images). Though McClintocks ideas were initially
considered sceptically, she later on received great honors including the Nobel Prize for Physiology and Medicine 1983 for her pioneering
work. Today, diverse types of transposable elements are described in almost all studied eukaryotic species. Transposable elements are typically
assorted into class 1 and class 2 transposons. While the former propagate via RNA intermediaries that are subject to reverse
transcription, class 2 transposons are excised from one locus and re-integrated at a different site via the enzyme
In
1950 Barbara McClintock first described mobile DNA elements in maize, where transposing DNA elements cause reversible disruptions of coloring
genes (right top: Science cover from 20. Nov. 2009, Photo: Kate Mathis/Getty Images). Though McClintocks ideas were initially
considered sceptically, she later on received great honors including the Nobel Prize for Physiology and Medicine 1983 for her pioneering
work. Today, diverse types of transposable elements are described in almost all studied eukaryotic species. Transposable elements are typically
assorted into class 1 and class 2 transposons. While the former propagate via RNA intermediaries that are subject to reverse
transcription, class 2 transposons are excised from one locus and re-integrated at a different site via the enzyme
 transposase. Mobile DNA can account for a considerable fraction of metazoan genomes. Conservative estimations assume that roughly 50% of a mammalian genome
are made up of repetitive DNA. Especially class 1 transposons of endogenous (e.g. the human specific 7SL RNA-derived Alu elements) or exogenous (e.g. ERV)
origin steadily increased in copy number by applying the copy-paste-like cloning mechanism for their own propagation throughout the host genome,
resulting in an increase of copy number with each transposition.
transposase. Mobile DNA can account for a considerable fraction of metazoan genomes. Conservative estimations assume that roughly 50% of a mammalian genome
are made up of repetitive DNA. Especially class 1 transposons of endogenous (e.g. the human specific 7SL RNA-derived Alu elements) or exogenous (e.g. ERV)
origin steadily increased in copy number by applying the copy-paste-like cloning mechanism for their own propagation throughout the host genome,
resulting in an increase of copy number with each transposition. Different classes of transposons are active in diverse animals and constitute a steady threat to genome integrity by potentially disrupting essential genes and providing the prerequisite for ectopic
recombination (right bottom: Active human class 1 transposons). Throughout the metazoan lineage, organisms have established a molecular defense mechanism that employes Piwi proteins
and a broad arsenal of small noncoding RNAs (Piwi-interacting RNAs, piRNAs) to track down hostile retroposon RNA-intermediates via RNA interference (RNAi). On the other
hand, new classes of retroposons arised and existing retroposons constantly diversified over evolutionary timespans thus eluding the established host resistence. Constant host adaptation results in a
molecular arms race between transposons and the Piwi/piRNA pathway components resulting in fast evolutionary changes driven by positive selection. Evolutionary
patterns become even more complex considering benefical functionalization events of transposon sequences through e.g. exonization, promotor capturing or miRNA processing from terminal
inverted repeats (TIRs) of class 2 elements.
Different classes of transposons are active in diverse animals and constitute a steady threat to genome integrity by potentially disrupting essential genes and providing the prerequisite for ectopic
recombination (right bottom: Active human class 1 transposons). Throughout the metazoan lineage, organisms have established a molecular defense mechanism that employes Piwi proteins
and a broad arsenal of small noncoding RNAs (Piwi-interacting RNAs, piRNAs) to track down hostile retroposon RNA-intermediates via RNA interference (RNAi). On the other
hand, new classes of retroposons arised and existing retroposons constantly diversified over evolutionary timespans thus eluding the established host resistence. Constant host adaptation results in a
molecular arms race between transposons and the Piwi/piRNA pathway components resulting in fast evolutionary changes driven by positive selection. Evolutionary
patterns become even more complex considering benefical functionalization events of transposon sequences through e.g. exonization, promotor capturing or miRNA processing from terminal
inverted repeats (TIRs) of class 2 elements.A non-representative selection of relevant literature
Biémont and Vieira 2006 Nature 443:521-524 PubMed link
Slotkin and Martienssen 2007 Nat Rev Genet 8:272-285 PubMed link
Hancks and Kazazian 2012 Cur Opin Genet Dev 22:191-203 PubMed link
Fedoroff 2012 Science 338:758-767 PubMed link
Lisch 2013 Nat Rev Genet 14:49-61 PubMed link
__________________________________________________________________________________________________________
__________________________________________________________________________________________________________

 Small noncoding RNAs (sncRNAs) represent a diverse class of regulatory RNAs ranging from 18-34 nt in length being involved in a variety of
biological processes. Bound to Argonaute proteins they act at the heart of RNA induced silencing complexes (RISCs) to regulate gene expression
or defend the genome from transposable elements on the transcriptional and post-transcriptional level.
Small noncoding RNAs (sncRNAs) represent a diverse class of regulatory RNAs ranging from 18-34 nt in length being involved in a variety of
biological processes. Bound to Argonaute proteins they act at the heart of RNA induced silencing complexes (RISCs) to regulate gene expression
or defend the genome from transposable elements on the transcriptional and post-transcriptional level.sncRNAs can be categorized into three main classes (further classes exist): micro (mi-) RNAs, short interferring (si-) RNAs and Piwi-interacting (pi-) RNAs. Both, miRNAs and siRNAs are products of Dicer processing, an enzyme that recognizes double stranded RNA, and bind to the Argonaute-like subclass of Argonaute proteins. In contrast, piRNAs are processed from single stranded transcripts either by Zucchini (primary piRNAs from piRNA cluster transcripts) or in a self sustaining amplification loop referred to as ping-pong-cycle involving typically two different paralogs of the Piwi-like subclass of Argonaute proteins that alternately slice transcripts from piRNA clusters and complementary target loci such as (but not exclusively) transposons (bottom: piRNA biogenesis. Figure from Luteijn and Ketting 2013. Nature Review Genetics 14:523-534 with reuse liscence #3595390394990).
 While miRNAs are involved in post-transcriptional gene regulation in both plants and animals, piRNAs and Piwi proteins (that are absent in
plants) have been linked to transciptional and post-transcriptional transposon control in the animal germline, though evidence for diverse functions beyond
transposon repression is mounting. In plants, transposons are controlled by siRNAs which also represent the major defense line against RNA viruses. In
contrast, animal siRNAs are involved in transposon controll and gene regulation though the significance of siRNAi in animals remains an
outstanding question and appears to be relegated to the background compared to miRNA- and piRNA-pathways.
While miRNAs are involved in post-transcriptional gene regulation in both plants and animals, piRNAs and Piwi proteins (that are absent in
plants) have been linked to transciptional and post-transcriptional transposon control in the animal germline, though evidence for diverse functions beyond
transposon repression is mounting. In plants, transposons are controlled by siRNAs which also represent the major defense line against RNA viruses. In
contrast, animal siRNAs are involved in transposon controll and gene regulation though the significance of siRNAi in animals remains an
outstanding question and appears to be relegated to the background compared to miRNA- and piRNA-pathways.
__________________________________________________________________________________________________________
- How secondary structure influences small RNA pathways
Transposable elements in the tree shrew are targeted not only by piRNAs but also by smaller RNAs resembling typical features of siRNAs. Comprehensive bioinformatic analyses showed that piRNAs preferentially target transposon transcripts at sites that do not form foldback structures and thus remain accessible for guiding piRNAs. In contrast, sites that avoid piRNA targeting owing to strong secundary structures are processed by Dicer, an enzyme that recognizes double-stranded RNA (Rosenkranz et al. 2015, RNA 21(5):911-922).

- How temperature influences small RNA pathways
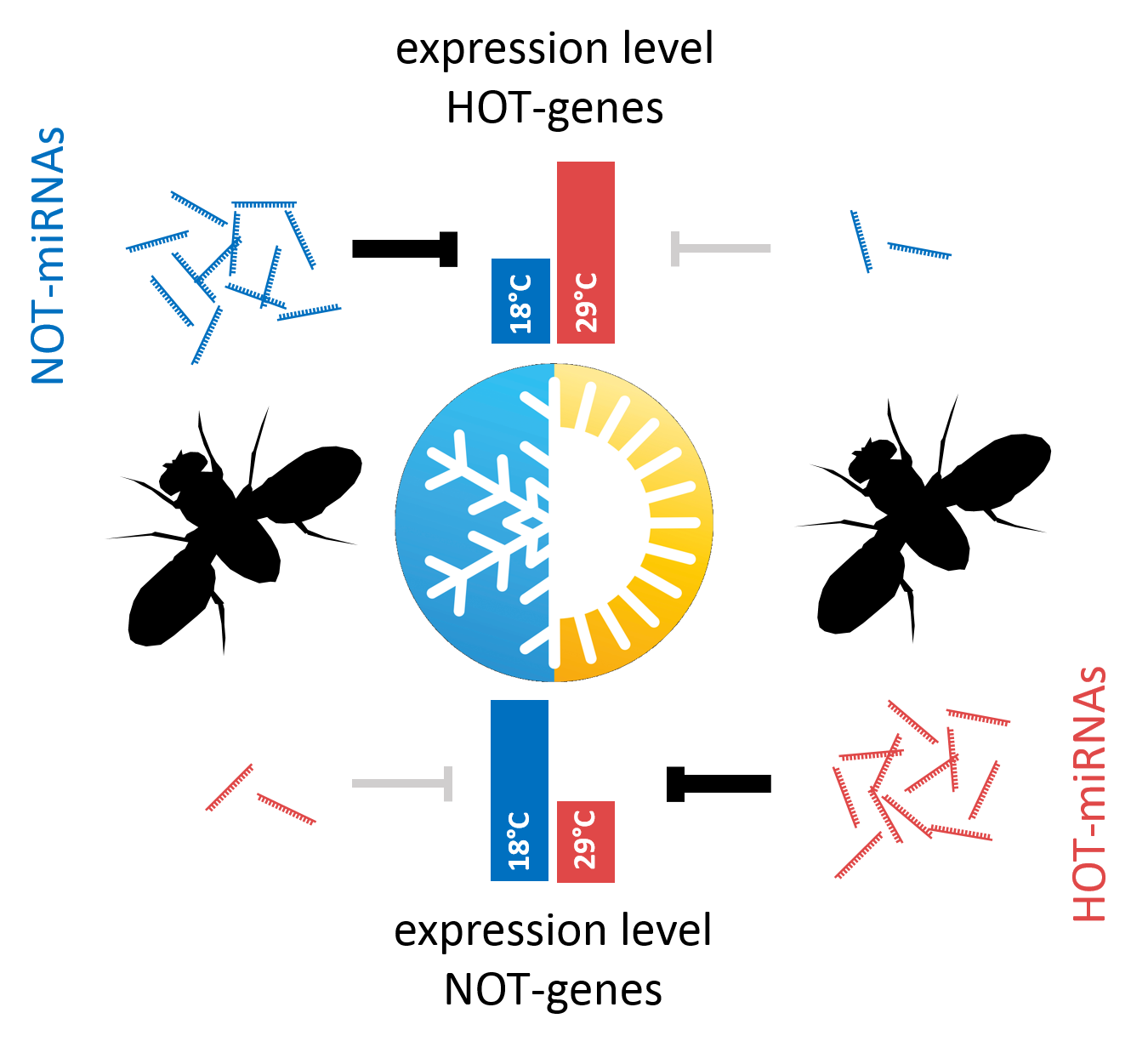
Altered ambient temperature induces drastic, but reversible changes in sequence composition and total abundance of both, miRNA- and piRNA populations in Drosophila. Regarding miRNAs, we can distinguish between miRNAs with higher abundance at Hyper-Optimal Temperature (29°C, HOT-miRNAs) and miRNAs with higher abundance at Near-Optimal Temperature (18°C, NOT-miRNAs). Further, the expression of temperature-responsive miRNAs and their predicted target transcripts correlates inversely, suggesting that temperature-responsive miRNAs drive adaptation to different ambient temperatures on the transcriptome level.
In addition, altered temperature not only affects miRNA expression but also influences piRNA biogenesis. We observed significantly increased ping-pong processing at 29°C which is presumably driven by dissolved RNA secondary structures at higher temperatures, uncovering target sites that are not accessible at low temperatures. (Fast et al. 2017, RNA 23(9):1352-1364).
- Near-neutral sequence evolution of piRNA clusters and piRNAs
piRNA clusters do not evolve uniformely. Instead, we can observe different conservation patterns within the same piRNA clusters. Residues that are overrepresented in tree shrew (Tupaia belangeri) testis piRNA transcriptomes show stronger sequence conservation, whereas piRNA coding residues in general show only very weak sequence conservation . In contrast, regulatory elements such as A-Myb transcription factor binding sites exhibit stronger sequence conservation, showing that the ability of a genomic locus to produce piRNAs is more important than preserving primary sequences of piRNAs (Rosenkranz et al. 2015, RNA 21(5):911-922).
Altered ambient temperature induces drastic, but reversible changes in sequence composition and total abundance of both, miRNA- and piRNA populations in Drosophila. Regarding miRNAs, we can distinguish between miRNAs with higher abundance at Hyper-Optimal Temperature (29°C, HOT-miRNAs) and miRNAs with higher abundance at Near-Optimal Temperature (18°C, NOT-miRNAs). Further, the expression of temperature-responsive miRNAs and their predicted target transcripts correlates inversely, suggesting that temperature-responsive miRNAs drive adaptation to different ambient temperatures on the transcriptome level.
In addition, altered temperature not only affects miRNA expression but also influences piRNA biogenesis. We observed significantly increased ping-pong processing at 29°C which is presumably driven by dissolved RNA secondary structures at higher temperatures, uncovering target sites that are not accessible at low temperatures. (Fast et al. 2017, RNA 23(9):1352-1364).

- Near-neutral sequence evolution of piRNA clusters and piRNAs
piRNA clusters do not evolve uniformely. Instead, we can observe different conservation patterns within the same piRNA clusters. Residues that are overrepresented in tree shrew (Tupaia belangeri) testis piRNA transcriptomes show stronger sequence conservation, whereas piRNA coding residues in general show only very weak sequence conservation . In contrast, regulatory elements such as A-Myb transcription factor binding sites exhibit stronger sequence conservation, showing that the ability of a genomic locus to produce piRNAs is more important than preserving primary sequences of piRNAs (Rosenkranz et al. 2015, RNA 21(5):911-922).

- Conservation of piRNA-mediated gene regulatory function
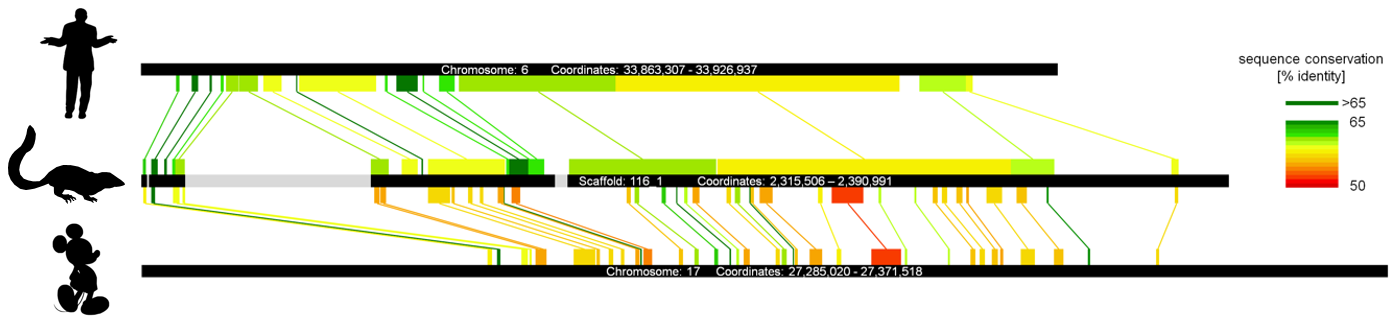
While in flys piRNAs are enriched for transposon sequences, the majority of mammalian pachytene piRNAs matches single-copy sequences. This raised the question of whether the Piwi/piRNA system is involved in functions beyond transposon silencing. Our in-deep characterization of the porcine testis piRNA transcriptome provided evidence for a processing of protein-coding transcripts within the piRNA ping-pong cycle. Remarkably, orthologous genes (but not simply highly expressed genes) are targeted by piRNAs accross different mammalian species pointing towards a conserved function of the Piwi/piRNA pathway in post-transcriptional gene regulation in mammals (Gebert et al. 2015, PLoS ONE 10(5):e0124860).
While in flys piRNAs are enriched for transposon sequences, the majority of mammalian pachytene piRNAs matches single-copy sequences. This raised the question of whether the Piwi/piRNA system is involved in functions beyond transposon silencing. Our in-deep characterization of the porcine testis piRNA transcriptome provided evidence for a processing of protein-coding transcripts within the piRNA ping-pong cycle. Remarkably, orthologous genes (but not simply highly expressed genes) are targeted by piRNAs accross different mammalian species pointing towards a conserved function of the Piwi/piRNA pathway in post-transcriptional gene regulation in mammals (Gebert et al. 2015, PLoS ONE 10(5):e0124860).