


Upcoming Events, Lectures and Talks
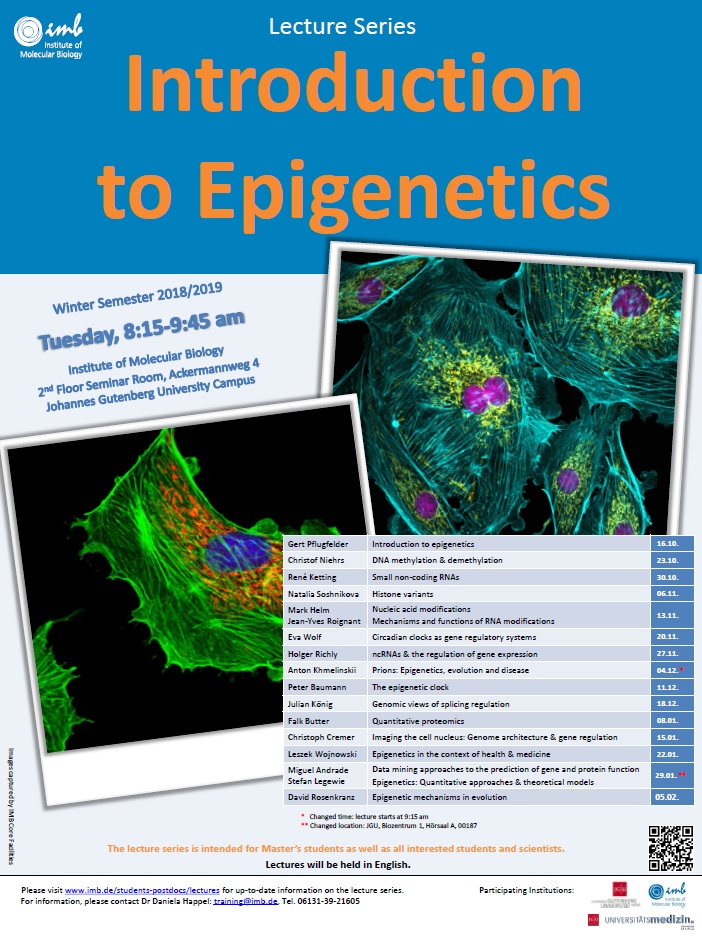 Lecture Series “Introduction to Epigenetics” for this year’s Winter Semester 2018/2019.
Lectures will take place on Tuesdays, 8:15-9:45 am at IMB and will start next Tuesday, 16 October.
Lecture Series “Introduction to Epigenetics” for this year’s Winter Semester 2018/2019.
Lectures will take place on Tuesdays, 8:15-9:45 am at IMB and will start next Tuesday, 16 October.
Soon...
Tuesday, 5th February 2019, 08:15: Introduction into Epigenetics - Epigenetic Mechanisms in Evolution / Recent work of the small RNA group. Institute of Molecular Biology (IMB) Mainz.
Friday, 8th February 2019, 08:15: Humane Epigenetik, Audimax (alte Mensa) Universitätscampus.
Information for students
PDF files from lectures and talks can be downloaded below. Files are compressed and decompression requires a password since some documents may contain unpublished data or third party material. The files are for personal use only. You will get the password at the end of each lecture (or contact David Rosenkranz).
IPP-ISS Blocklecture summer 2015: Epigenetic mechanisms in Evolution
IPP-ISS Blocklecture winter 2015/16: piRNAs and Piwi proteins: function and evolution
Master Anthropology 2016: Lab course sRNA extraction/cloning/sequencing/analysis sequence data
Master Anthropology lecture summer 2016: Epigenetic mechanisms in Evolution
IPP-ISS Blocklecture summer 2016 (1+2): Epigenetic mechanisms in Evolution / piRNAs and Piwi proteins
IPP-ISS Blocklecture winter 2016/17: piRNAs and Piwi proteins: function and evolution
Master Anthropology. Module 3 - Lab course summer 2017
IPP-ISS Blocklecture summer 2017 (1+2): Epigenetic mechanisms in Evolution / HOT or NOT
Literature for 'Modul14A' winter 2017/18
'Humane Epigenetik, Vorlesung aus der Reihe Humanbiologie und Anthropologie' winter 2017/18
'Epigenetics/Evolution (Modul 3)', summer 2018
IPP-ISS Blocklecture summer 2018 (1+2): Epigenetic mechanisms in Evolution / HOT or NOT
Introduction to Epigenetics 2019 (1+2): Epigenetic mechanisms in Evolution / Projects of the small RNA group Mainz
'Humane Epigenetik, Vorlesung aus der Reihe Humanbiologie und Anthropologie' winter 2018/19
Master students may also want to access our Extended Lab Protocols for the 'Master Anthropology' courses.
Applications for a Bachelor-, Master- or PhD position are welcome!
Interested and motivated bachelor/master/PhD students are welcome to apply for a thesis position in our group. We currently offer a PhD project in the IPP summer call 2016. The candidates are expected to have a good biomolecular background. Beside the wet lab work the projects will imply bioinformatic analyses. Skills in a programming language such as Perl, Python or C/C++ are advantageous. At least we expect the candidates to aquire these skills in the course of their thesis.
The candidates will learn and apply a number of classical molecular lab techniques such as:
- DNA/RNA Extraction and Purification
- PCR and quantitative Real-Time PCR (qPCR)
- Agarose/Polyacrilamide Gelelectrophoresis
- Cell culture techniques incl. knock-down experiments
- Co-Immunoprecipitation (Co-IP)
- Western Blot
- NGS Library Preparation
- Molecular Cloning
- Sanger Sequencing
Many projects include the analysis of Next Generation Sequencing data. The extent of data produced by these new sequencing techniques requires robust automated processing and analysis methods. Therefore the candidates have to apply bioinformatic tools that for the most part represent in-house Perl scripts that have been developed in our group. However, analyses will likely require the customization of existing, or the development of new Perl scripts. Large scale analysis can be performed on the institute's high performance linux server (64 cores, 260 GB RAM).
Below you find some recommendations for further reading.







 Lecture Series “Introduction to Epigenetics” for this year’s Winter Semester 2018/2019.
Lectures will take place on Tuesdays, 8:15-9:45 am at IMB and will start next Tuesday, 16 October.
Lecture Series “Introduction to Epigenetics” for this year’s Winter Semester 2018/2019.
Lectures will take place on Tuesdays, 8:15-9:45 am at IMB and will start next Tuesday, 16 October.
Soon...
Tuesday, 5th February 2019, 08:15: Introduction into Epigenetics - Epigenetic Mechanisms in Evolution / Recent work of the small RNA group. Institute of Molecular Biology (IMB) Mainz.
Friday, 8th February 2019, 08:15: Humane Epigenetik, Audimax (alte Mensa) Universitätscampus.
Information for students
PDF files from lectures and talks can be downloaded below. Files are compressed and decompression requires a password since some documents may contain unpublished data or third party material. The files are for personal use only. You will get the password at the end of each lecture (or contact David Rosenkranz).
IPP-ISS Blocklecture summer 2015: Epigenetic mechanisms in Evolution
IPP-ISS Blocklecture winter 2015/16: piRNAs and Piwi proteins: function and evolution
Master Anthropology 2016: Lab course sRNA extraction/cloning/sequencing/analysis sequence data
Master Anthropology lecture summer 2016: Epigenetic mechanisms in Evolution
IPP-ISS Blocklecture summer 2016 (1+2): Epigenetic mechanisms in Evolution / piRNAs and Piwi proteins
IPP-ISS Blocklecture winter 2016/17: piRNAs and Piwi proteins: function and evolution
Master Anthropology. Module 3 - Lab course summer 2017
IPP-ISS Blocklecture summer 2017 (1+2): Epigenetic mechanisms in Evolution / HOT or NOT
Literature for 'Modul14A' winter 2017/18
'Humane Epigenetik, Vorlesung aus der Reihe Humanbiologie und Anthropologie' winter 2017/18
'Epigenetics/Evolution (Modul 3)', summer 2018
IPP-ISS Blocklecture summer 2018 (1+2): Epigenetic mechanisms in Evolution / HOT or NOT
Introduction to Epigenetics 2019 (1+2): Epigenetic mechanisms in Evolution / Projects of the small RNA group Mainz
'Humane Epigenetik, Vorlesung aus der Reihe Humanbiologie und Anthropologie' winter 2018/19
Master students may also want to access our Extended Lab Protocols for the 'Master Anthropology' courses.
Applications for a Bachelor-, Master- or PhD position are welcome!
Interested and motivated bachelor/master/PhD students are welcome to apply for a thesis position in our group. We currently offer a PhD project in the IPP summer call 2016. The candidates are expected to have a good biomolecular background. Beside the wet lab work the projects will imply bioinformatic analyses. Skills in a programming language such as Perl, Python or C/C++ are advantageous. At least we expect the candidates to aquire these skills in the course of their thesis.
The candidates will learn and apply a number of classical molecular lab techniques such as:
- DNA/RNA Extraction and Purification
- PCR and quantitative Real-Time PCR (qPCR)
- Agarose/Polyacrilamide Gelelectrophoresis
- Cell culture techniques incl. knock-down experiments
- Co-Immunoprecipitation (Co-IP)
- Western Blot
- NGS Library Preparation
- Molecular Cloning
- Sanger Sequencing
Many projects include the analysis of Next Generation Sequencing data. The extent of data produced by these new sequencing techniques requires robust automated processing and analysis methods. Therefore the candidates have to apply bioinformatic tools that for the most part represent in-house Perl scripts that have been developed in our group. However, analyses will likely require the customization of existing, or the development of new Perl scripts. Large scale analysis can be performed on the institute's high performance linux server (64 cores, 260 GB RAM).
Below you find some recommendations for further reading.



















 1
1 2
2 3
3 4
4





